New prognostic tool detects “undetectable” tumor DNA in bloodstream
One way to determine how successfully a patient’s cancer treatment has eradicated the disease is to check the bloodstream for free-floating DNA originating from tumor cells, also known as circulating tumor DNA (ctDNA). The detection of ctDNA can serve as a powerful prognostic tool, allowing clinicians to assess the effectiveness of treatment and predict the likelihood of disease recurrence. Current ctDNA detection methods are limited, however, and may produce “false negatives.” Take, for example, patients with diffuse large B-cell lymphoma (DLBCL), the most common form of non-Hodgkin lymphoma, who can expect a favorable outcome if their ctDNA is below a certain level after two cycles of therapy. But nearly a third of DLBCL patients who eventually experience relapse do not have ctDNA detected in their bloodstream at this time – meaning we need a stronger magnifying glass.
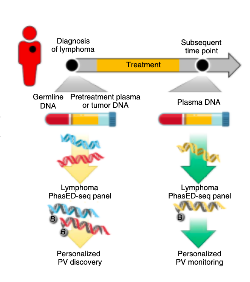
to identify phased variants
At Stanford University, Damon Runyon alumni Ash Alizadeh, MD, PhD, and David Kurtz, MD, PhD, have developed a screening method that, remarkably, was able to detect ctDNA in one out of every four DLBCL patients whose ctDNA had been pronounced undetectable after two cycles of therapy. Their new approach builds upon the current one, wherein free-floating (or “cell-free”) DNA is screened for mutations that match those of the tumor genome. To minimize background noise, recent methods use “duplex sequencing,” meaning the mutation must appear on both strands of DNA in order to get flagged. This makes sense in theory, but in practice, it is not always possible to recover both strands from the patient’s blood sample. More false negatives abound.
With a stroke of innovation, Dr. Kurtz and Dr. Alizadeh have found a way around this problem. Their approach relies on “phased variants,” where two or more mutations occur on the same strand of DNA. To develop the screen, the researchers identified regions in the lymphoid cancer genome where these mutations occur no further apart than the length of a typical cell-free DNA fragment. Known as phased variant enrichment and detection sequencing (PhasED-seq), this approach still requires multiple mutations to confirm the detection of ctDNA, but the results are not compromised by partial DNA recovery.
As ctDNA detection gains popularity as a prognostic tool, improved detection methods could make a world of difference for patients undergoing cancer therapy. Those without detectable ctDNA by PhasED-seq after treatment may be able to abbreviate their chemotherapy course; those who would have received a “false negative” from previous methods, but are in fact likely to experience relapse, may benefit from additional treatment. With clinical trials now on the horizon, PhasED-seq shows promise as the eagle-eyed lens we have been missing.
This research was published in Nature Biotechnology.
